Features
Github Repository
Forum
About Us
☰
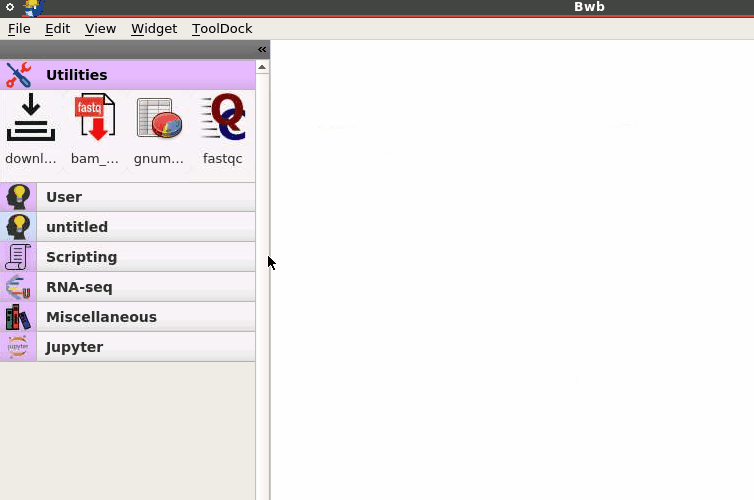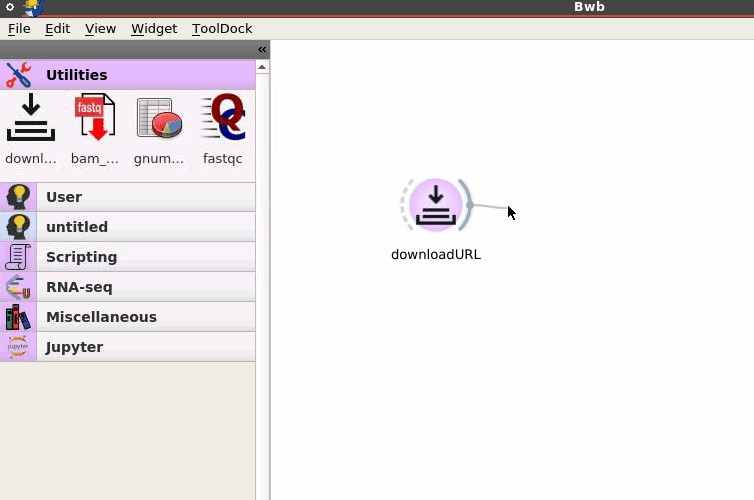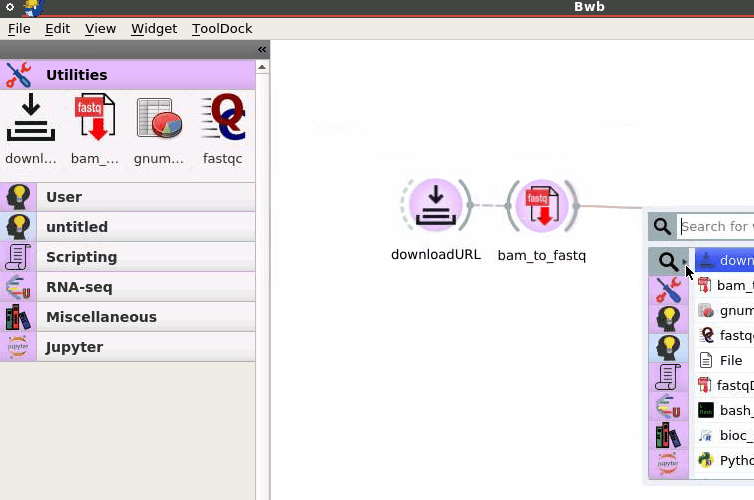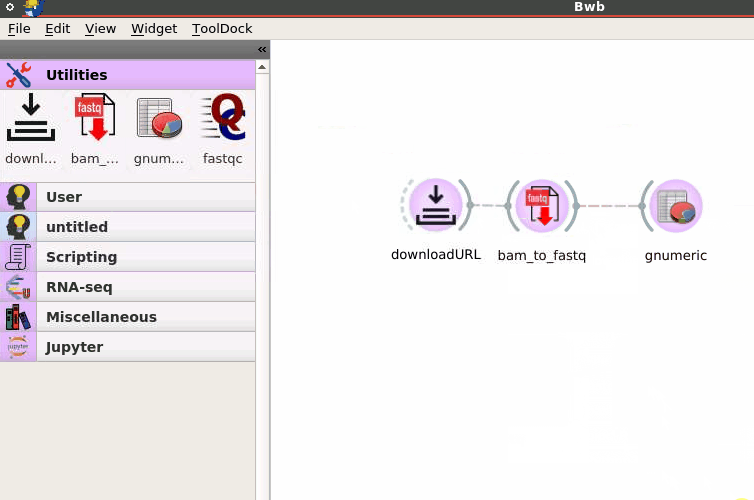
Users can create and execute modular bioinformatics workflows using a drag-and-drop interface. The Bwb GUI allows a non-programmer to install, customize, and reproducibly apply a workflow to their data with minimum effort.

Bwb provides a form-based user interface to facilitate parameter entry and a console to display intermediate results. New modules can be added without writing code. Created workflows can easily be exported and shared as a shell script or Bwb's native's format.

Bwb workflows can be run locally or on the cloud. Our use of Docker containers ensures that results and environments are always consistent.